Error when generating fragment libraries
- mostafa.razavi
-
 Topic Author
Topic Author
- Offline
- Junior Member
-

Less
More
- Posts: 22
- Thank you received: 1
5 years 5 months ago #550
by mostafa.razavi
Dear Cassandra community,
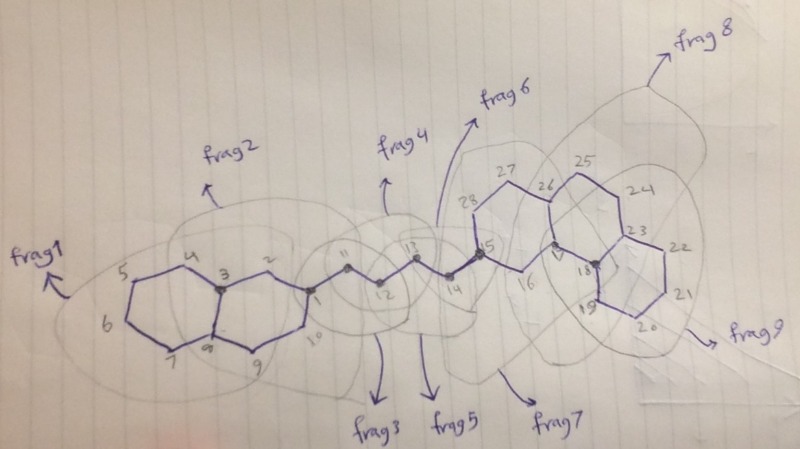
I'm trying to run NVT simulation of a rather complicated molecule that has 5 rings. The molecule is 1-naphthalenyl,4-phenanthrenyl butane. I'm attaching a drawing of the molecule with numbered united-atom sites as well a zip file containing ff, mcf, pdb, and input file. When I run the following command to generate the fragment libraries. I get an error. Can anyone help me figure out what I'm doing wrong? Also, I noticed that some atoms (number 3, 17, and 18) in Atom_Info section of mcf file have two "rings", while other ring atoms have only one "rings". Is it because they are part of two rings AND they are the center of the fragment they belong to?
Thank you,
Mostafa Razavi
I'm trying to run NVT simulation of a rather complicated molecule that has 5 rings. The molecule is 1-naphthalenyl,4-phenanthrenyl butane. I'm attaching a drawing of the molecule with numbered united-atom sites as well a zip file containing ff, mcf, pdb, and input file. When I run the following command to generate the fragment libraries. I get an error. Can anyone help me figure out what I'm doing wrong? Also, I noticed that some atoms (number 3, 17, and 18) in Atom_Info section of mcf file have two "rings", while other ring atoms have only one "rings". Is it because they are part of two rings AND they are the center of the fragment they belong to?
$ python2.7 library_setup.py cassandra_gfortran.exe nvt.inp 1-p-4-n-B.pdb
********************** Cassandra Setup *********************
Cassandra location: /home/mostafa/Git/Cassandra_V1.2/Src/cassandra_gfortran.exe
Scanning input file
Number of species found: 1
The MCF file number 1 is: 1-p-4-n-B.mcf
Species 1 has 9 fragments
Molecules with rings found. These are:
Species 1 has 5 rings.
Creating input MCF generation file for species 1
Running Cassandra to generate MCF files
One of the local ids is zero
This occurred while writing the angle info section
for the ring fragment mcf file
Fragment 2
Species 1
This error occurred in subroutine Write_Ring_Fragment_MCF_Angle_Info on step 0.
Fatal Error. Stopping program.
Traceback (most recent call last):
File "/home/mostafa/Git/Cassandra_V1.2/Scripts/Frag_Library_Setup/library_setup.py", line 619, in <module>
angle_type = line.split()[4]Thank you,
Mostafa Razavi
Please Log in to join the conversation.
- jshah46545
-

- Offline
- Junior Member
-

Less
More
- Posts: 27
- Thank you received: 5
5 years 5 months ago #551
by jshah46545
Mostafa,
As the script for generating fragment does not currently support fused rings, it incorrectly identifies that the molecule contains 9 fragments. The molecule has the following fragments (any (fused) ring is considered as a fragment along with its exoring atoms) :
Fragment 1: atoms 1-11
Fragment 2: atoms 1, 11, 12
Fragment 3: 11, 12, 13
Fragment 4: 12, 13, 14
Fragment 5: atoms 14-28
If you manually modify the # Fragment_Info and # Fragment_Connectivity sections in the mcf file and rerun the fragment generation script, it might work.
Please let us know if you continue to have issues.
Best,
Jindal
As the script for generating fragment does not currently support fused rings, it incorrectly identifies that the molecule contains 9 fragments. The molecule has the following fragments (any (fused) ring is considered as a fragment along with its exoring atoms) :
Fragment 1: atoms 1-11
Fragment 2: atoms 1, 11, 12
Fragment 3: 11, 12, 13
Fragment 4: 12, 13, 14
Fragment 5: atoms 14-28
If you manually modify the # Fragment_Info and # Fragment_Connectivity sections in the mcf file and rerun the fragment generation script, it might work.
Please let us know if you continue to have issues.
Best,
Jindal
Please Log in to join the conversation.
- mostafa.razavi
-
 Topic Author
Topic Author
- Offline
- Junior Member
-

Less
More
- Posts: 22
- Thank you received: 1
5 years 5 months ago #552
by mostafa.razavi
Jindal,
Thank you for your helpful response. I manually specified Fragment_Info and Fragment_Connectivity sections in mcf file as below:
The script ran successfully, but Cassandra gave an error:
Then I tried considering 13, 14, 15 as a fragment, with the same logic as we consider 1, 11, 12 a fragment. The script and Cassandra both run successfully:
Is the above specification correct? Specifically, have I identified the branch_point values for the first and last fragments correctly?
Thank you for your helpful response. I manually specified Fragment_Info and Fragment_Connectivity sections in mcf file as below:
!index number_of_atoms_in_fragment branch_point other_atoms
# Fragment_Info
5
1 11 1 2 3 4 5 6 7 8 9 10 11
2 3 11 1 12
3 3 12 11 13
4 3 13 12 14
5 15 15 16 17 18 19 20 21 22 23 24 25 26 27 28 14
# Fragment_Connectivity
4
1 1 2
2 2 3
3 3 4
4 4 5The script ran successfully, but Cassandra gave an error:
Number of atoms in species 1 is 28
Number of atoms deleted with frag 1 is 9
Number of atoms deleted with frag 2 is 4
This error occurred in subroutine Get_Fragment_Connectivity_Info on step 0.
Fatal Error. Stopping program.Then I tried considering 13, 14, 15 as a fragment, with the same logic as we consider 1, 11, 12 a fragment. The script and Cassandra both run successfully:
!index number_of_atoms_in_fragment branch_point other_atoms
# Fragment_Info
6
1 11 1 2 3 4 5 6 7 8 9 10 11
2 3 11 1 12
3 3 12 11 13
4 3 13 12 14
5 3 14 13 15
6 15 15 16 17 18 19 20 21 22 23 24 25 26 27 28 14
# Fragment_Connectivity
5
1 1 2
2 2 3
3 3 4
4 4 5
5 5 6Is the above specification correct? Specifically, have I identified the branch_point values for the first and last fragments correctly?
Please Log in to join the conversation.
- mostafa.razavi
-
 Topic Author
Topic Author
- Offline
- Junior Member
-

Less
More
- Posts: 22
- Thank you received: 1
5 years 5 months ago #553
by mostafa.razavi
Also, I realized that in the mcf file generated by mcfgen.py script, atoms 3, 17, and 18 have "ring ring" in the Atom_Info section. I cannot think of a special trait that these atoms might have, other than being part of two rings. If this is true, should 8, 23, and 26 also have "ring ring" instead of "ring"?
# Atom_Info
28
1 CA Ca 12.01070 0.0 LJ 21.000 3.880 ring
2 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
3 C1A 1A 12.01070 0.0 LJ 30.000 3.700 ring ring
4 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
5 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
6 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
7 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
8 C1A 1A 12.01070 0.0 LJ 30.000 3.700 ring
9 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
10 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
11 CH2 H2 14.02660 0.0 LJ 46.000 3.950
12 CH2 H2 14.02660 0.0 LJ 46.000 3.950
13 CH2 H2 14.02660 0.0 LJ 46.000 3.950
14 CH2 H2 14.02660 0.0 LJ 46.000 3.950
15 CA Ca 12.01070 0.0 LJ 21.000 3.880 ring
16 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
17 C1A 1A 12.01070 0.0 LJ 30.000 3.700 ring ring
18 C1A 1A 12.01070 0.0 LJ 30.000 3.700 ring ring
19 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
20 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
21 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
22 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
23 C1A 1A 12.01070 0.0 LJ 30.000 3.700 ring
24 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
25 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
26 C1A 1A 12.01070 0.0 LJ 30.000 3.700 ring
27 CH Ch 13.01864 0.0 LJ 50.500 3.695 ring
28 CH Ch 13.01864 0.0 LJ 50.500 3.695 ringPlease Log in to join the conversation.
- ryangmullen
-

- Offline
- Administrator
-

Less
More
- Posts: 124
- Karma: 4
- Thank you received: 24
5 years 5 months ago #554
by ryangmullen
Hi Mostafa,
Yes, atoms 13, 14, 15 should be identified as their own fragment and everything looks correct in your # Fragment_Info and # Fragment_Connectivity sections.
I think "ring ring" and "ring" will both be read the same by Cassandra. Are you treating fragments 1 and 6 (the fused ring fragments) as flexible or rigid?
Ryan
Yes, atoms 13, 14, 15 should be identified as their own fragment and everything looks correct in your # Fragment_Info and # Fragment_Connectivity sections.
I think "ring ring" and "ring" will both be read the same by Cassandra. Are you treating fragments 1 and 6 (the fused ring fragments) as flexible or rigid?
Ryan
Please Log in to join the conversation.
- mostafa.razavi
-
 Topic Author
Topic Author
- Offline
- Junior Member
-

Less
More
- Posts: 22
- Thank you received: 1
5 years 5 months ago #555
by mostafa.razavi
Thanks Ryan,
I'm simulating this molecule using TraPPE-UA force field, so yes I'm trying to treat the fused rings as rigid. I set all the bonds, and angles to be fixed, but I have dihedrals as you can see in the mcf file.
I'm simulating this molecule using TraPPE-UA force field, so yes I'm trying to treat the fused rings as rigid. I set all the bonds, and angles to be fixed, but I have dihedrals as you can see in the mcf file.
Please Log in to join the conversation.
Time to create page: 0.185 seconds