Unable to generate the fragment molecules
- wthor1
-
 Topic Author
Topic Author
- Offline
- New Member
-

Less
More
- Posts: 2
- Thank you received: 0
5 years 1 month ago #588
by wthor1
Dear all,
Recently I've tried to use cassandra on my system, it should be a polymer, and I've some questions on it.
When I'm trying to generate the fragments using library_setup.py, it generated the error message as shown:
----
Creating input MCF generation file for species 1
Running Cassandra to generate MCF files
Error attempting to parse line 562 of input file:
2 22 6 1 2 3 4 5 23 24 26 28 36 53 7
into at least 24 entries
This error occurred in subroutine Parse_String on step 0.
Fatal Error. Stopping program.
Note: The following floating-point exceptions are signalling: IEEE_DENORMAL
Traceback (most recent call last):
File "library_setup.py", line 604, in <module>
"_1.mcf",'r')
IOError: [Errno 2] No such file or directory: 'species1/fragments/frag_1_1.mcf'
----
I've generated the mcf files using mcfgen.py, and this error occurs.
I was wondering is there any upper limits for the fragments atomic info?
Thank you in advance!
Recently I've tried to use cassandra on my system, it should be a polymer, and I've some questions on it.
When I'm trying to generate the fragments using library_setup.py, it generated the error message as shown:
----
Creating input MCF generation file for species 1
Running Cassandra to generate MCF files
Error attempting to parse line 562 of input file:
2 22 6 1 2 3 4 5 23 24 26 28 36 53 7
into at least 24 entries
This error occurred in subroutine Parse_String on step 0.
Fatal Error. Stopping program.
Note: The following floating-point exceptions are signalling: IEEE_DENORMAL
Traceback (most recent call last):
File "library_setup.py", line 604, in <module>
"_1.mcf",'r')
IOError: [Errno 2] No such file or directory: 'species1/fragments/frag_1_1.mcf'
----
I've generated the mcf files using mcfgen.py, and this error occurs.
I was wondering is there any upper limits for the fragments atomic info?
Thank you in advance!
Please Log in to join the conversation.
- emarin
-

- Offline
- Administrator
-

Less
More
- Posts: 42
- Karma: 2
- Thank you received: 7
- wthor1
-
 Topic Author
Topic Author
- Offline
- New Member
-

Less
More
- Posts: 2
- Thank you received: 0
5 years 1 month ago #592
by wthor1
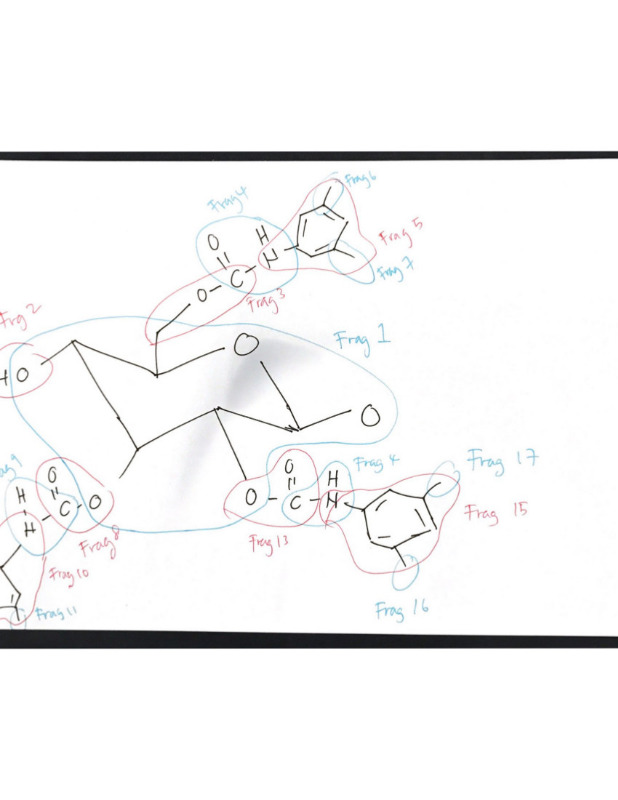
When I check the fragment info from the mcfgen.py, maybe there are quit many rings in my system (I am using monomer at the moment), so the fragments connectivity are a little messy, I decide to redefine the fragments and it is as shown as image below:
After that, I did not get the error as shown above, and at least 2 fragments are generated (better than last time, nothing generated?), however, I get error like this:
"""
Scanning input file
Number of species found: 1
The MCF file number 1 is: pilot_ATM.mcf
Species 1 has 17 fragments
Molecules with rings found. These are:
Species 1 has 5 rings.
Creating input MCF generation file for species 1
Running Cassandra to generate MCF files
Program received signal SIGSEGV: Segmentation fault - invalid memory reference.
Backtrace for this error:
#0 0x7FE304F47E08
#1 0x7FE304F46F90
#2 0x7FE3046784AF
#3 0x489BFB in participation_
#4 0x4783E6 in mcf_control_
#5 0x409D86 in MAIN__ at main.f90:?
Traceback (most recent call last):
File "library_setup.py", line 604, in <module>
"_1.mcf",'r')
IOError: [Errno 2] No such file or directory: 'species1/fragments/frag_3_1.mcf'
"""
I rechecked my structure and the connectivity, I guess it should be fine so I've no idea what's the mistake I've done here.
Any suggestion or correction please?
Much appreciated and have a good day!
I attach my mcf file also for better reference.
After that, I did not get the error as shown above, and at least 2 fragments are generated (better than last time, nothing generated?), however, I get error like this:
"""
Scanning input file
Number of species found: 1
The MCF file number 1 is: pilot_ATM.mcf
Species 1 has 17 fragments
Molecules with rings found. These are:
Species 1 has 5 rings.
Creating input MCF generation file for species 1
Running Cassandra to generate MCF files
Program received signal SIGSEGV: Segmentation fault - invalid memory reference.
Backtrace for this error:
#0 0x7FE304F47E08
#1 0x7FE304F46F90
#2 0x7FE3046784AF
#3 0x489BFB in participation_
#4 0x4783E6 in mcf_control_
#5 0x409D86 in MAIN__ at main.f90:?
Traceback (most recent call last):
File "library_setup.py", line 604, in <module>
"_1.mcf",'r')
IOError: [Errno 2] No such file or directory: 'species1/fragments/frag_3_1.mcf'
"""
I rechecked my structure and the connectivity, I guess it should be fine so I've no idea what's the mistake I've done here.
Any suggestion or correction please?
Much appreciated and have a good day!
I attach my mcf file also for better reference.
Please Log in to join the conversation.
Time to create page: 0.171 seconds